
Grant R. Nickles, PhD
UW Distinguished Research Fellow
Wisconsin Research, Innovation and Scholarly Excellence (RISE) AI
College of Agriculture and Life Sciences
at The University of Wisconsin-Madison
Research Projects
Exploring the evolution and genomic architecture of non-canonical fungal specialized metabolism
I am interested in studying non-canonical fungal specialized metabolism which broadly encompasses: (i) natural products whose genetic basis is poorly understood or unpredictable by conventional genome mining, and (ii) taxa who have been conventionally understudied or ignored by natural product researchers (e.g., Basidiomycetes, lichens).
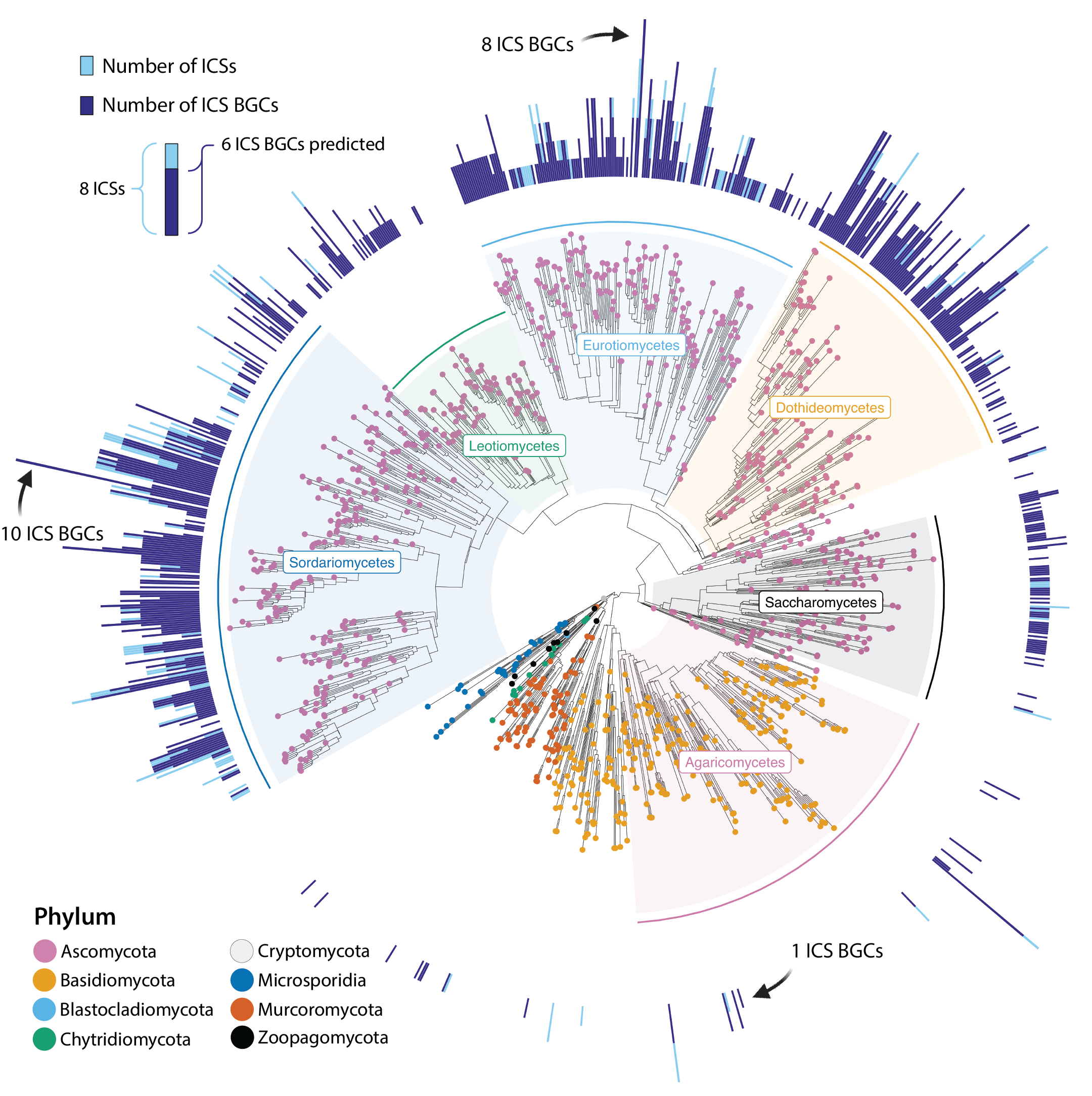
Mining for a new class of fungal natural products
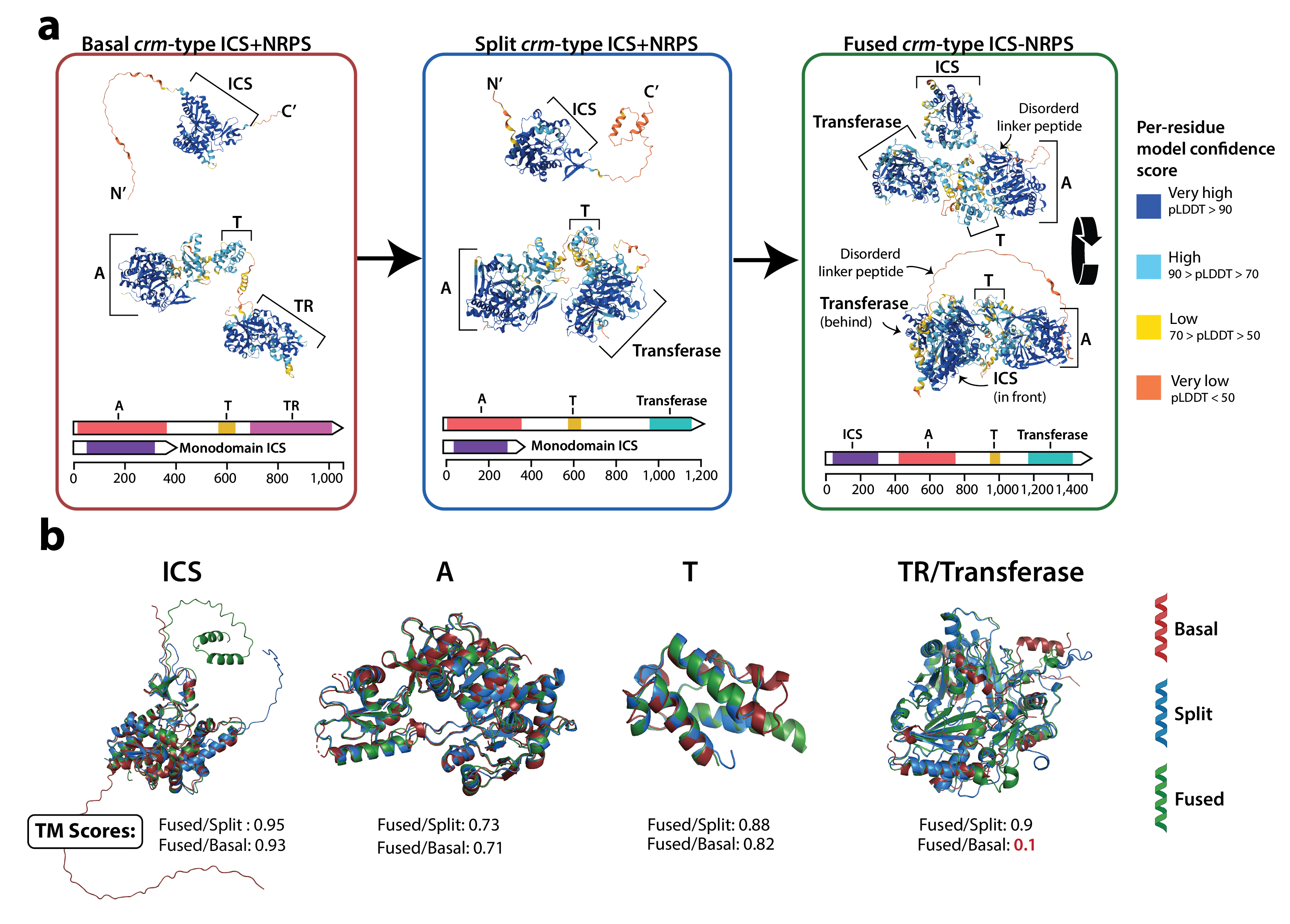
Unconvering the stepwise evolution of a fungal mega-synthase with the help of lichen genomes
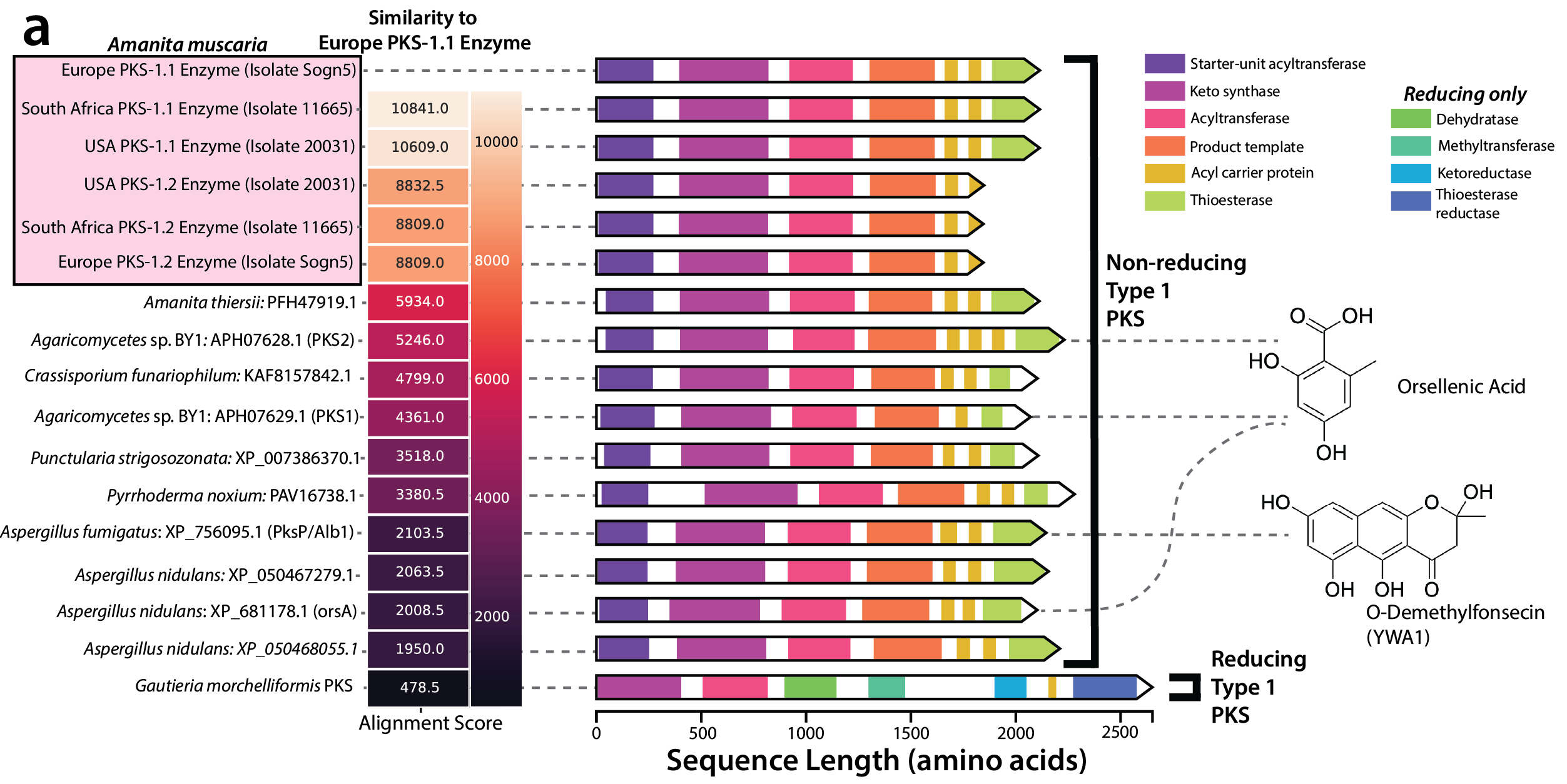
The invasion of Amanita muscaria in South Africa

Aspergillus fumigatus Af293/CEA10 Community Sequencing Initiative
The model pathogen A. fumigatus (strains Af293 and CEA10) has been used by researchers for decades. As these strains were cultured independently in labs across the globe, they have inevitably accumulated unique, unstudied mutations. This "lab-to-lab" evolution makes it difficult to compare findings on physiology, pathogenicity, and genetics.
While community sequencing projects are common for other model organisms (like Arabidopsis, S. cerevisiae, and various bacteria), this effort is the first of its kind for a filamentous fungus. We believe A. fumigatus is the perfect model pathogen to pioneer this work. This is a global collaboration that we hope will provide a foundational genomic resource for the entire Aspergillus community for years to come.
I launched the Aspergillus fumigatus Community Sequencing Initiative in collaboration with Dr. Amelia Barber, Dr. Emile Gluck-Thaler, and Dr. Dante Calise. We founded this project on the principle that reliable, comparable data is crucial for scientific communities.
Creating tools to study the metabolisms of mixed microbial communities
The core aim of my work is to expand our understanding of the chemical interplay that occurs in mixed (fungal and bacterial) microbial communities. Toward this, I am developing two parallel approaches: running large-scale in silico protein docking experiments to identify candidate enzymes, and generating new, stable model fungal-bacterial communities whose collective metabolomes can be studied in a lab setting.
I believe any viable microbial bioremediation strategy will ultimately involve the collective catabolic capabilities of many microbes working in concert. I hope to later build on the foundations of my model mixed microbial communities toward engineering consortia that can target and degrade specific chemicals.
Contact & CV
I'm always interested in discussing collaborations on fungal natural product biosynthesis and evolution. Feel free to reach out!
Get in Touch
-
Email
gnickles@wisc.edu
gnick317@gmail.com -
Connect with me
- LinkedInLinkedIn
- GitHubGitHub
- Research GateResearchGate
- OrcIDOrcID
- 🦋Bluesky
-
Publications & Citations
View my complete publication list and citation metrics on Google Scholar.
View Google Scholar Profile -
Scientific Community Memberships
Member of the Genetics Society of America (GSA)